| Ⅰ | This article along with all titles and tags are the original content of AppNee. All rights reserved. To repost or reproduce, you must add an explicit footnote along with the URL to this article! |
| Ⅱ | Any manual or automated whole-website collecting/crawling behaviors are strictly prohibited. |
| Ⅲ | Any resources shared on AppNee are limited to personal study and research only, any form of commercial behaviors are strictly prohibited. Otherwise, you may receive a variety of copyright complaints and have to deal with them by yourself. |
| Ⅳ | Before using (especially downloading) any resources shared by AppNee, please first go to read our F.A.Q. page more or less. Otherwise, please bear all the consequences by yourself. |
| This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. |
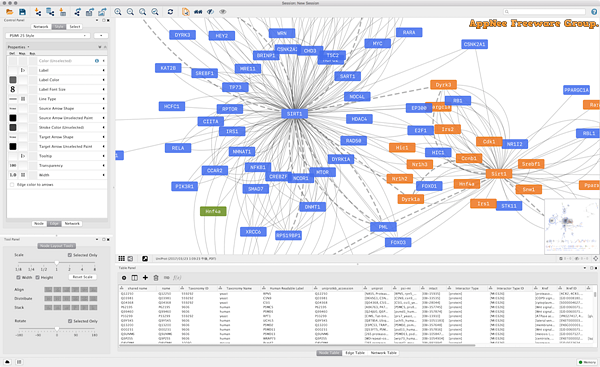
If you are a biology student, researcher, or simply a biology enthusiast eager to deepen your understanding, Cytoscape is the ideal tool for you. This software enables you to visualize molecular interaction networks and biological pathways while incorporating annotations and state data.
Cytoscape is a free and open-source software platform widely utilized for visualizing complex networks and integrating these networks with any type of attribute data. Initially developed for the biological research community, particularly in the fields of genomics and proteomics, it has expanded its applications to various domains, including social network analysis, systems biology, and even information systems.
Cytoscape is able to create visually intuitive graphs that represent relationships and interactions among different biological entities, such as genes, proteins, and metabolites. With some completed projects, you’ll gain a better grasp of its capabilities. It includes a network management panel, a network viewing window, and an attribute browser, allowing you to easily modify various attributes.
You can load multiple networks simultaneously, analyze them, and make adjustments as needed. However, there may be instances where certain networks are not viewable due to their size, which can delay display times. It can assist you in identifying which networks are viewable by using black and red highlights to indicate their status. For large networks that are viewable, you can examine their structure from a bird’s-eye perspective, allowing you to pan across the network to see its nodes and zoom in for detailed or comprehensive views.
Cytoscape provides valuable support for systems biology, genomics, and proteomics. It allows you to import protein-protein interaction datasets in various formats, including Graph Markup Language, SBML, BioPAX, GraphML, and Excel Workbook. Additionally, its capabilities extend beyond biology, as it can also process and analyze extensive social networks and visualize Resource Description Framework data.
Overall, Cytoscape is a sophisticated application designed to handle both simple and complex data for visualizing and analyzing interaction networks in both biological and social science domains.
// Key Features //
| Feature | Description |
| Supports Many Standards | Cytoscape supports a lot of standard network and annotation file formats including: SIF (Simple Interaction Format), GML, XGMML, BioPAX, PSI-MI, GraphML, KGML (KEGG XML), SBML, OBO, and Gene Association. Delimited text files and MS Excel™ Workbook are also supported and you can import data files, such as expression profiles or GO annotations, generated by other applications or spreadsheet programs. Using this feature, you can load and save arbitrary attributes on nodes, edges, and networks. For example, input a set of custom annotation terms for your proteins, create a set of confidence values for your protein-protein interactions. |
| Public Database Clients | Cytoscape works as a web service client. This means Cytoscape can directly connect to the external public databases and imports network and annotation data. Currently, Pathway Commons, IntAct, BioMart, and NCBI Entrez Gene are supported. And we continue to develop new service clients for popular databases. |
| Interoperability | Since Cytoscape supports import/export standard file formats, you can easily put Cytoscape into your workflow. For example, if you have a network data generated by igraph or Bioconductor, Cytoscape can load the file as a text table and you can export it in PSI-MI format for other bioinformatics tools or your own applications/scripts.
From version 3.3, Cytoscape supports RESTful API for programmatic access. You can use your choice of programming languages to access Cytoscape core features, such as creating networks, applying layouts, or image export. |
| Session File | You can save your work by a single click. All of settings, data files, and visualizations are packed in a session file. It is called Cytoscape Session (.cys) file. Cytoscape Session file includes networks, attributes (for node/edge/network), Desktop states (selected/hidden nodes and edges, window sizes), Properties, some plugin states, and Visual Styles. |
| Layout | Layout networks in two dimensions. A variety of layout algorithms are available, including cyclic, tree, force-directed, edge-weight, and yFiles Organic layouts. You can also use Manual Layout tools similar to other graphics application user interface. |
| Image Export | You can export networks as publishable-quality images. Supports PDF, PS, SVG, PNG, JPEG, and BMP files. Vector images (PDF and PS) can be modified by other application such as Adobe Illustrator for further enhancements. |
| VizMapper | Customize network data display using powerful VisualStyles. View a superposition of gene expression ratios and p-values on the network. Expression data can be mapped to node color, label, border thickness, or border color, etc. according to user-configurable colors and visualization schemes. |
| Filter | Filter the network to select subsets of nodes and/or interactions based on the current data. For instance, users may select nodes involved in a threshold number of interactions, nodes that share a particular GO annotation, or nodes whose gene expression levels change significantly in one or more conditions according to p-values loaded with the gene expression data. You can create new networks from the filtering result. |
| Search | Search target nodes and edges from Search window. Lucene Syntax is supported for arbitrary complex queries. |
| Browsing | Zoom in/out and pan for browsing the network. Use the network manager to easily organize multiple networks. And this structure can be saved in a session file. Use the bird’s eye view to easily navigate large networks. Easily navigate large networks (100,000+ nodes and edges) by efficient rendering engine. |
| Find Modules/ Clusters | Find active subnetworks/pathway modules. The network is screened against gene expression data to identify connected sets of interactions, i.e. interaction subnetworks, whose genes show particularly high levels of differential expression. The interactions contained in each subnetwork provide hypotheses for the regulatory and signaling interactions in control of the observed expression changes. Find clusters (highly interconnected regions) in any network loaded into Cytoscape. Depending on the type of network, clusters may mean different things. For instance, clusters in a protein-protein interaction network have been shown to be protein complexes and parts of pathways. Clusters in a protein similarity network represent protein families. |
| App Manager and App Store | Apps are available for network and molecular profile analysis. Cytoscape is a software written in Java and you can write your own App for data analysis, import, and visualization by writing Java code. More Apps are available at the Cytoscape App Store. You can install most of the Apps by just one click from App Manager, or directly install from the App Store (this is a Cytoscape 3 feature). |
| Export to Web | Cytoscape 3 can generate a web application using Cytoscape.js from your Cytoscape session. In addtion, you can easily export your networks as JSON files, and share with your collaborators using external web application such as cyNetShare. |
| Multilingual | You can use your language for the data files. Most of the features in Cytoscape supports languages other than English, including Eastern Asian languages using two-byte characters. |
// Main Applications //
| Feature | Description |
| Biology | Cytoscape supports many use cases in molecular and systems biology, genomics, and proteomics:
|
| Social Science | Cytoscape is used by social scientists to:
|
| General Complex Network Analysis | Cytoscape is domain-independent and therefore is a powerful tool for complex network analysis in general.
|
| App Development | Cytoscape is expandable and extensible. Cytoscape has a vibrant App developer community and over a hundred Apps developed by third parties.
|
// Official Demo Video //
// Related Links //
// Download URLs //
| License | Version | Download | Size |
| Freeware | Latest |  (mir) (mir) |
n/a |
| If some download link is missing, and you do need it, just please send an email (along with post link and missing link) to remind us to reupload the missing file for you. And, give us some time to respond. | |
| If there is a password for an archive, it should be "appnee.com". | |
| Most of the reserved downloads (including the 32-bit version) can be requested to reupload via email. |